Runs a Bayesian meta-analysis assuming that the mean effect \(d\) in each study is identical (i.e., a fixed-effects analysis).
Arguments
- y
effect size per study. Can be provided as (1) a numeric vector, (2) the quoted or unquoted name of the variable in
data, or (3) aformulato include discrete or continuous moderator variables.- SE
standard error of effect size for each study. Can be a numeric vector or the quoted or unquoted name of the variable in
data- labels
optional: character values with study labels. Can be a character vector or the quoted or unquoted name of the variable in
data- data
data frame containing the variables for effect size
y, standard errorSE,labels, and moderators per study.- d
priordistribution on the average effect sized. The prior probability density function is defined viaprior.- rscale_contin
scale parameter of the JZS prior for the continuous covariates.
- rscale_discrete
scale parameter of the JZS prior for discrete moderators.
- centering
whether continuous moderators are centered.
- logml
how to estimate the log-marginal likelihood: either by numerical integration (
"integrate") or by bridge sampling using MCMC/Stan samples ("stan"). To obtain high precision withlogml="stan", many MCMC samples are required (e.g.,logml_iter=10000, warmup=1000).- summarize
how to estimate parameter summaries (mean, median, SD, etc.): Either by numerical integration (
summarize = "integrate") or based on MCMC/Stan samples (summarize = "stan").- ci
probability for the credibility/highest-density intervals.
- rel.tol
relative tolerance used for numerical integration using
integrate. Userel.tol=.Machine$double.epsfor maximal precision (however, this might be slow).- silent_stan
whether to suppress the Stan progress bar.
- ...
further arguments passed to
rstan::sampling(seestanmodel-method-sampling). Relevant MCMC settings concern the number of warmup samples that are discarded (warmup=500), the total number of iterations per chain (iter=2000), the number of MCMC chains (chains=4), whether multiple cores should be used (cores=4), and control arguments that make the sampling in Stan more robust, for instance:control=list(adapt_delta=.97).
Examples
### Bayesian Fixed-Effects Meta-Analysis (H1: d>0)
data(towels)
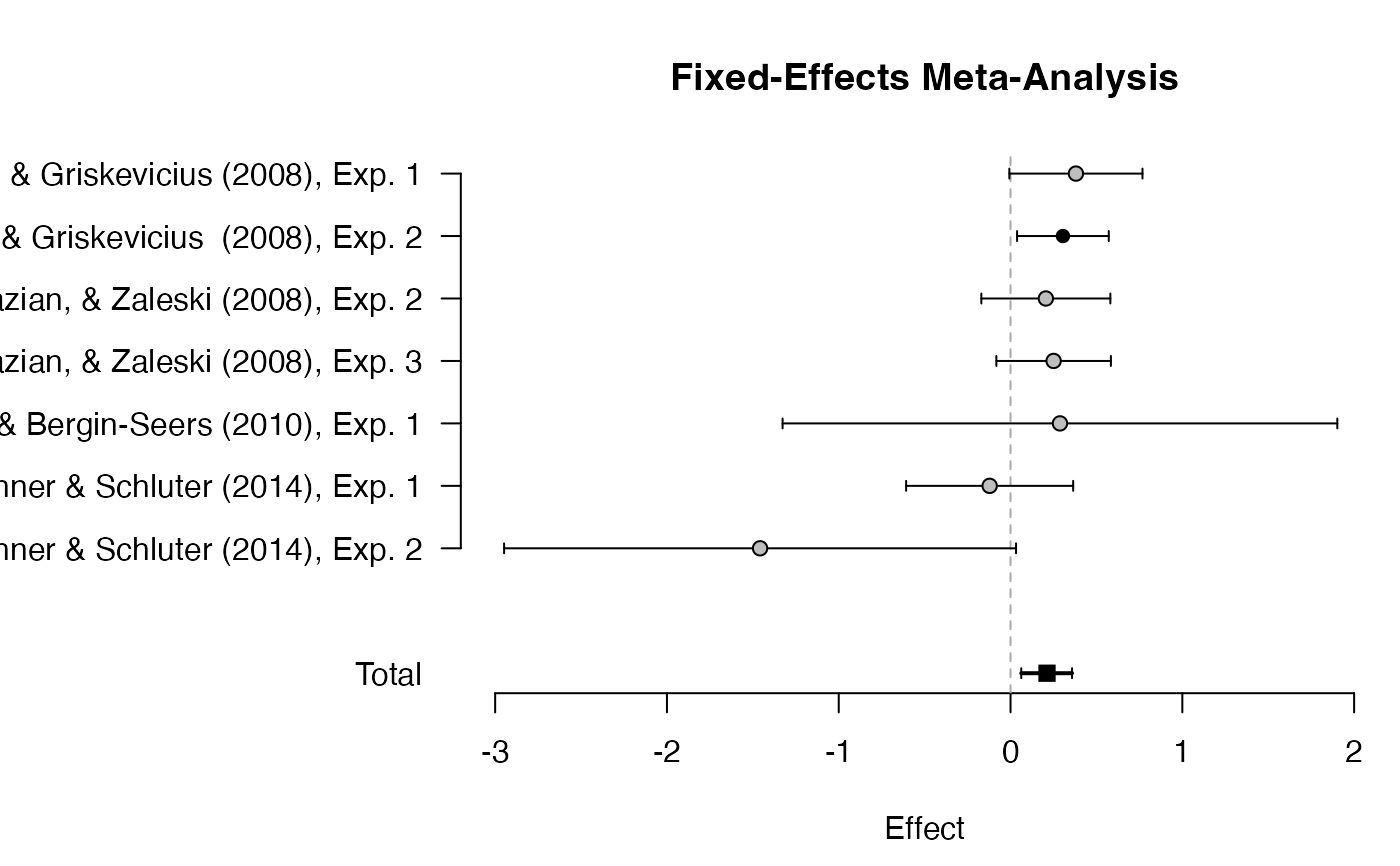
mf <- meta_fixed(logOR, SE, study,
data = towels,
d = prior("norm", c(mean = 0, sd = .3), lower = 0)
)
mf
#> ### Bayesian Fixed-Effects Meta-Analysis ###
#> Prior on d: 'norm' (mean=0, sd=0.3) truncated to the interval [0,Inf].
#>
#> # Bayes factors:
#> (denominator)
#> (numerator) fixed_H0 fixed_H1
#> fixed_H0 1.0 0.0419
#> fixed_H1 23.9 1.0000
#>
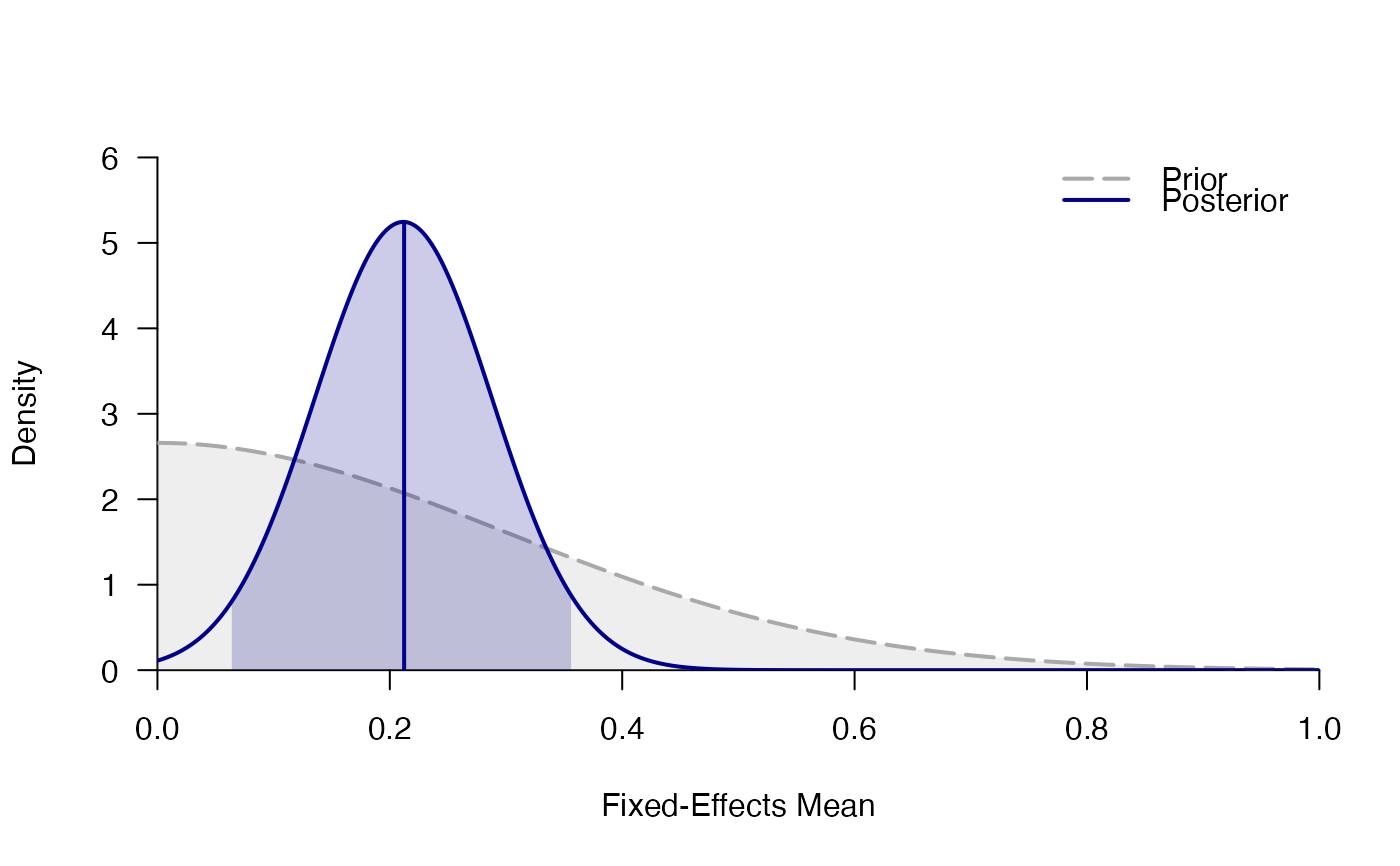
#> # Posterior summary statistics of fixed-effects model:
#> mean sd 2.5% 50% 97.5% hpd95_lower hpd95_upper n_eff Rhat
#> d 0.212 0.075 0.066 0.212 0.361 0.062 0.358 NA NA
plot_posterior(mf)
 plot_forest(mf)
plot_forest(mf)