Fits random- and fixed-effects meta-analyses and performs Bayesian model averaging for H1 (d != 0) vs. H0 (d = 0).
meta_bma(
y,
SE,
labels,
data,
d = prior("cauchy", c(location = 0, scale = 0.707)),
tau = prior("invgamma", c(shape = 1, scale = 0.15)),
rscale_contin = 0.5,
rscale_discrete = 0.707,
centering = TRUE,
prior = c(1, 1, 1, 1),
logml = "integrate",
summarize = "stan",
ci = 0.95,
rel.tol = .Machine$double.eps^0.3,
logml_iter = 5000,
silent_stan = TRUE,
...
)Arguments
- y
effect size per study. Can be provided as (1) a numeric vector, (2) the quoted or unquoted name of the variable in
data, or (3) aformulato include discrete or continuous moderator variables.- SE
standard error of effect size for each study. Can be a numeric vector or the quoted or unquoted name of the variable in
data- labels
optional: character values with study labels. Can be a character vector or the quoted or unquoted name of the variable in
data- data
data frame containing the variables for effect size
y, standard errorSE,labels, and moderators per study.- d
priordistribution on the average effect sized. The prior probability density function is defined viaprior.- tau
priordistribution on the between-study heterogeneitytau(i.e., the standard deviation of the study effect sizesdstudyin a random-effects meta-analysis. A (nonnegative) prior probability density function is defined viaprior.- rscale_contin
scale parameter of the JZS prior for the continuous covariates.
- rscale_discrete
scale parameter of the JZS prior for discrete moderators.
- centering
whether continuous moderators are centered.
- prior
prior probabilities over models (possibly unnormalized) in the order
c(fixed_H0, fixed_H1, random_H0, random_H1). For instance, if we expect fixed effects to be two times as likely as random effects and H0 and H1 to be equally likely:prior = c(2,2,1,1).- logml
how to estimate the log-marginal likelihood: either by numerical integration (
"integrate") or by bridge sampling using MCMC/Stan samples ("stan"). To obtain high precision withlogml="stan", many MCMC samples are required (e.g.,logml_iter=10000, warmup=1000).- summarize
how to estimate parameter summaries (mean, median, SD, etc.): Either by numerical integration (
summarize = "integrate") or based on MCMC/Stan samples (summarize = "stan").- ci
probability for the credibility/highest-density intervals.
- rel.tol
relative tolerance used for numerical integration using
integrate. Userel.tol=.Machine$double.epsfor maximal precision (however, this might be slow).- logml_iter
number of iterations (per chain) from the posterior distribution of
dandtau. The samples are used for computing the marginal likelihood of the random-effects model with bridge sampling (iflogml="stan") and for obtaining parameter estimates (ifsummarize="stan"). Note that the argumentiter=2000controls the number of iterations for estimation of the random-effect parameters per study in random-effects meta-analysis.- silent_stan
whether to suppress the Stan progress bar.
- ...
further arguments passed to
rstan::sampling(seestanmodel-method-sampling). Relevant MCMC settings concern the number of warmup samples that are discarded (warmup=500), the total number of iterations per chain (iter=2000), the number of MCMC chains (chains=4), whether multiple cores should be used (cores=4), and control arguments that make the sampling in Stan more robust, for instance:control=list(adapt_delta=.97).
Details
Bayesian model averaging for four meta-analysis models: Fixed- vs. random-effects and H0 (\(d=0\)) vs. H1 (e.g., \(d>0\)). For a primer on Bayesian model-averaged meta-analysis, see Gronau, Heck, Berkhout, Haaf, and Wagenmakers (2020).
By default, the log-marginal likelihood is computed by numerical integration
(logml="integrate"). This is relatively fast and gives precise,
reproducible results. However, for extreme priors or data (e.g., very small
standard errors), numerical integration is not robust and might provide
incorrect results. As an alternative, the log-marginal likelihood can be
estimated using MCMC/Stan samples and bridge sampling (logml="stan").
To obtain posterior summary statistics for the average effect size d
and the heterogeneity parameter tau, one can also choose between
numerical integration (summarize="integrate") or MCMC sampling in Stan
(summarize="stan"). If any moderators are included in a model, both
the marginal likelihood and posterior summary statistics can only be computed
using Stan.
References
Gronau, Q. F., Erp, S. V., Heck, D. W., Cesario, J., Jonas, K. J., & Wagenmakers, E.-J. (2017). A Bayesian model-averaged meta-analysis of the power pose effect with informed and default priors: the case of felt power. Comprehensive Results in Social Psychology, 2(1), 123-138. doi:10.1080/23743603.2017.1326760
Gronau, Q. F., Heck, D. W., Berkhout, S. W., Haaf, J. M., & Wagenmakers, E.-J. (2021). A primer on Bayesian model-averaged meta-analysis. Advances in Methods and Practices in Psychological Science, 4(3), 1–19. doi:10.1177/25152459211031256
Berkhout, S. W., Haaf, J. M., Gronau, Q. F., Heck, D. W., & Wagenmakers, E.-J. (2023). A tutorial on Bayesian model-averaged meta-analysis in JASP. Behavior Research Methods.
See also
Examples
# \donttest{
### Bayesian Model-Averaged Meta-Analysis (H1: d>0)
data(towels)
set.seed(123)
mb <- meta_bma(logOR, SE, study, towels,
d = prior("norm", c(mean = 0, sd = .3), lower = 0),
tau = prior("invgamma", c(shape = 1, scale = 0.15))
)
mb
#> ### Meta-Analysis with Bayesian Model Averaging ###
#> Fixed H0: d = 0
#> Fixed H1: d ~ 'norm' (mean=0, sd=0.3) truncated to the interval [0,Inf].
#> Random H0: d = 0,
#> tau ~ 'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf].
#> Random H1: d ~ 'norm' (mean=0, sd=0.3) truncated to the interval [0,Inf].
#> tau ~ 'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf].
#>
#> # Bayes factors:
#> (denominator)
#> (numerator) fixed_H0 fixed_H1 random_H0 random_H1
#> fixed_H0 1.00 0.0419 0.372 0.0975
#> fixed_H1 23.87 1.0000 8.872 2.3259
#> random_H0 2.69 0.1127 1.000 0.2622
#> random_H1 10.26 0.4299 3.815 1.0000
#>
#> # Bayesian Model Averaging
#> Comparison: (fixed_H1 & random_H1) vs. (fixed_H0 & random_H0)
#> Inclusion Bayes factor: 9.249
#> Inclusion posterior probability: 0.902
#>
#> # Model posterior probabilities:
#> prior posterior logml
#> fixed_H0 0.25 0.0264 -5.58
#> fixed_H1 0.25 0.6311 -2.40
#> random_H0 0.25 0.0711 -4.59
#> random_H1 0.25 0.2713 -3.25
#>
#> # Posterior summary statistics of average effect size:
#> mean sd 2.5% 50% 97.5% hpd95_lower hpd95_upper n_eff Rhat
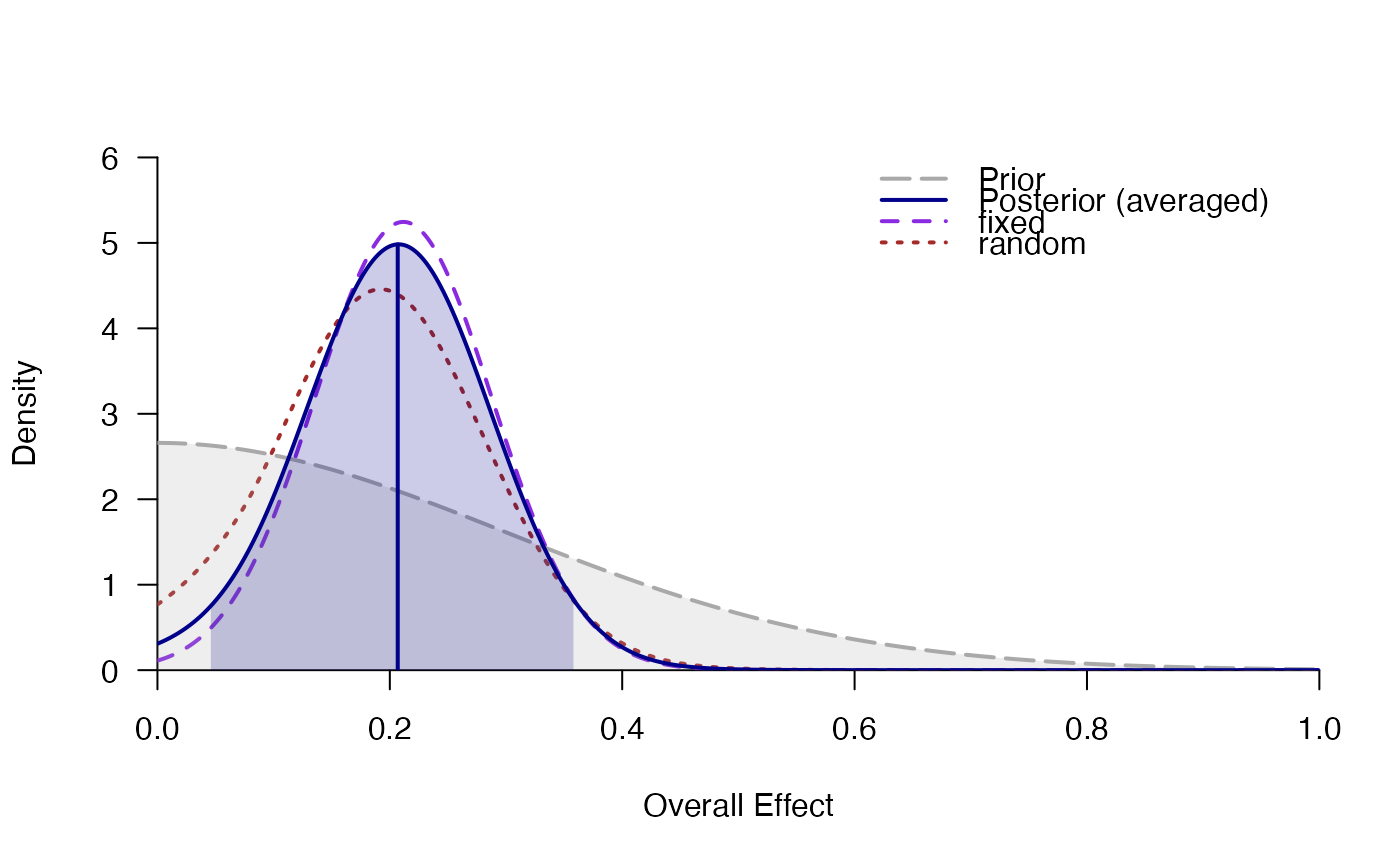
#> averaged 0.206 0.081 0.045 0.207 0.364 0.040 0.360 NA NA
#> fixed 0.214 0.075 0.070 0.215 0.364 0.073 0.365 2025.6 1.001
#> random 0.192 0.089 0.022 0.191 0.371 0.013 0.355 4846.4 1.001
plot_posterior(mb, "d")
 # }
# }