Sensitivity analysis assuming different prior distributions for the two main parameters of a Bayesian meta-analysis (i.e., the overall effect and the heterogeneity of effect sizes across studies).
meta_sensitivity(
y,
SE,
labels,
data,
d_list,
tau_list,
analysis = "bma",
combine_priors = "crossed",
...
)Arguments
- y
effect size per study. Can be provided as (1) a numeric vector, (2) the quoted or unquoted name of the variable in
data, or (3) aformulato include discrete or continuous moderator variables.- SE
standard error of effect size for each study. Can be a numeric vector or the quoted or unquoted name of the variable in
data- labels
optional: character values with study labels. Can be a character vector or the quoted or unquoted name of the variable in
data- data
data frame containing the variables for effect size
y, standard errorSE,labels, and moderators per study.- d_list
a
listof prior distributions specified viaprior()for the overall effect size (mean) across studies- tau_list
a
listof prior distributions specified viaprior()for the heterogeneity (SD) of effect sizes across studies- analysis
which type of meta-analysis should be performed for analysis? Can be one of the following:
"fixed"for fixed-effects model, seemeta_fixed()"random"for random-effects model, seemeta_random()"bma"for model averaging, seemeta_bma()
- combine_priors
either
"matched", in which case the analysis includes the matched pairwise combinations of the prior distributions specified ind_listandtau_list, orcrossed, in which case the analysis uses all possible pairwise combinations of priors.- ...
further arguments passed to the function specified in
analysis.
Value
an object of the S3 class meta_sensitivity, that is, a list of fitted
meta-analysis models. Results can be printed or plotted using
plot.meta_sensitivity().
See also
Examples
# \donttest{
data(towels)
sensitivity <- meta_sensitivity(
y = logOR, SE = SE, labels = study, data = towels,
d_list = list(prior("cauchy", c(0, .707)),
prior("norm", c(0, .5)),
prior("norm", c(.5, .3))),
tau_list = list(prior("invgamma", c(1, 0.15), label = "tau"),
prior("gamma", c(1.5, 3), label = "tau")),
analysis = "random",
combine_priors = "crossed")
#> Warning: There were 1 divergent transitions after warmup. See
#> https://mc-stan.org/misc/warnings.html#divergent-transitions-after-warmup
#> to find out why this is a problem and how to eliminate them.
#> Warning: Examine the pairs() plot to diagnose sampling problems
#> Warning: There were 2 divergent transitions after warmup. See
#> https://mc-stan.org/misc/warnings.html#divergent-transitions-after-warmup
#> to find out why this is a problem and how to eliminate them.
#> Warning: Examine the pairs() plot to diagnose sampling problems
#> Warning: There were 2 divergent transitions after warmup. See
#> https://mc-stan.org/misc/warnings.html#divergent-transitions-after-warmup
#> to find out why this is a problem and how to eliminate them.
#> Warning: Examine the pairs() plot to diagnose sampling problems
print(sensitivity, digits = 2)
#> ### Sensitivity analysis for Bayesian meta-analysis ###
#>
#> Prior distributions on d (= overall effect size):
#>
#> [1] "'t' (location=0, scale=0.707, nu=1) with support on the interval [-Inf,Inf]."
#> [2] "'t' (location=0, scale=0.707, nu=1) with support on the interval [-Inf,Inf]."
#> [3] "'norm' (mean=0, sd=0.5) with support on the interval [-Inf,Inf]."
#> [4] "'norm' (mean=0, sd=0.5) with support on the interval [-Inf,Inf]."
#> [5] "'norm' (mean=0.5, sd=0.3) with support on the interval [-Inf,Inf]."
#> [6] "'norm' (mean=0.5, sd=0.3) with support on the interval [-Inf,Inf]."
#>
#> Prior distributions on tau (= SD of effect sizes):
#>
#> [1] "'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf]."
#> [2] "'gamma' (shape=1.5, rate=3) with support on the interval [0,Inf]."
#> [3] "'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf]."
#> [4] "'gamma' (shape=1.5, rate=3) with support on the interval [0,Inf]."
#> [5] "'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf]."
#> [6] "'gamma' (shape=1.5, rate=3) with support on the interval [0,Inf]."
#>
#> Parameter estimates:
#>
#> prior mean sd 2.5% 50% 97.5% hpd95_lower hpd95_upper n_eff Rhat
#> d1 1 0.20 0.105 -0.026 0.20 0.39 -0.01344 0.40 4873 1
#> d2 2 0.19 0.124 -0.090 0.19 0.41 -0.06496 0.43 3602 1
#> d3 3 0.19 0.109 -0.036 0.20 0.39 -0.02826 0.40 4590 1
#> d4 4 0.18 0.127 -0.111 0.19 0.40 -0.07553 0.43 3511 1
#> d5 5 0.24 0.096 0.045 0.24 0.42 0.03746 0.42 5990 1
#> d6 6 0.24 0.108 0.024 0.24 0.45 0.02630 0.45 4448 1
#> tau1 1 0.13 0.103 0.033 0.11 0.38 0.02041 0.31 2397 1
#> tau2 2 0.18 0.141 0.015 0.14 0.54 0.00158 0.45 3166 1
#> tau3 3 0.14 0.117 0.033 0.11 0.41 0.01776 0.33 722 1
#> tau4 4 0.18 0.145 0.016 0.14 0.56 0.00053 0.46 3046 1
#> tau5 5 0.12 0.087 0.033 0.10 0.36 0.02086 0.29 4611 1
#> tau6 6 0.17 0.134 0.013 0.13 0.52 0.00035 0.43 3461 1
#>
#> Posterior model probabilities:
#>
#> random_H0 random_H1
#> prior1 0.55 0.45
#> prior2 0.63 0.37
#> prior3 0.41 0.59
#> prior4 0.50 0.50
#> prior5 0.38 0.62
#> prior6 0.47 0.53
par(mfrow = c(1,2))
plot(sensitivity, "d", "prior")
#> Prior distributions on d (= overall effect size):
#>
#> [1] "'t' (location=0, scale=0.707, nu=1) with support on the interval [-Inf,Inf]."
#> [2] "'t' (location=0, scale=0.707, nu=1) with support on the interval [-Inf,Inf]."
#> [3] "'norm' (mean=0, sd=0.5) with support on the interval [-Inf,Inf]."
#> [4] "'norm' (mean=0, sd=0.5) with support on the interval [-Inf,Inf]."
#> [5] "'norm' (mean=0.5, sd=0.3) with support on the interval [-Inf,Inf]."
#> [6] "'norm' (mean=0.5, sd=0.3) with support on the interval [-Inf,Inf]."
#>
#> Prior distributions on tau (= SD of effect sizes):
#>
#> [1] "'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf]."
#> [2] "'gamma' (shape=1.5, rate=3) with support on the interval [0,Inf]."
#> [3] "'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf]."
#> [4] "'gamma' (shape=1.5, rate=3) with support on the interval [0,Inf]."
#> [5] "'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf]."
#> [6] "'gamma' (shape=1.5, rate=3) with support on the interval [0,Inf]."
plot(sensitivity, "d", "posterior")
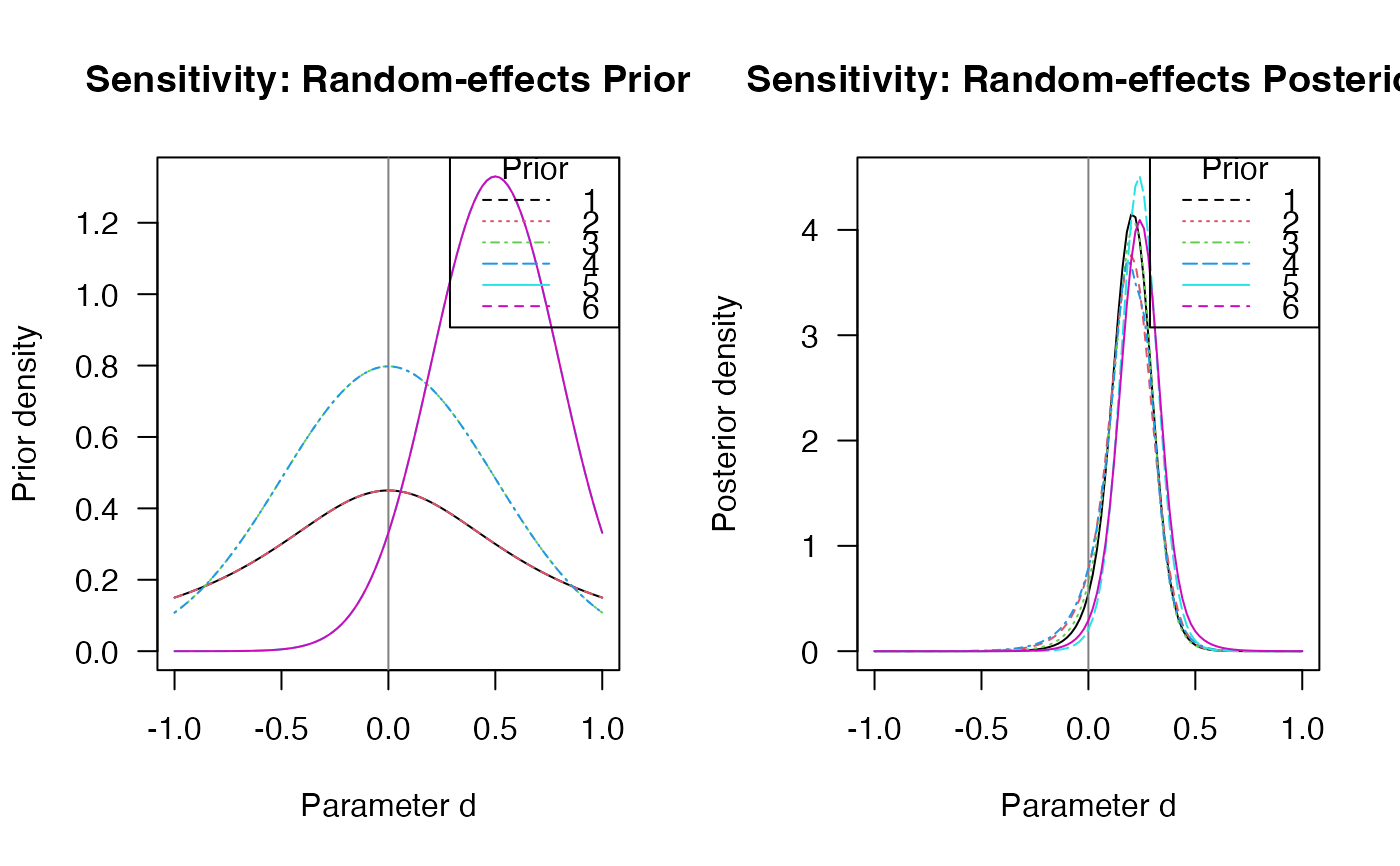 #> Prior distributions on d (= overall effect size):
#>
#> [1] "'t' (location=0, scale=0.707, nu=1) with support on the interval [-Inf,Inf]."
#> [2] "'t' (location=0, scale=0.707, nu=1) with support on the interval [-Inf,Inf]."
#> [3] "'norm' (mean=0, sd=0.5) with support on the interval [-Inf,Inf]."
#> [4] "'norm' (mean=0, sd=0.5) with support on the interval [-Inf,Inf]."
#> [5] "'norm' (mean=0.5, sd=0.3) with support on the interval [-Inf,Inf]."
#> [6] "'norm' (mean=0.5, sd=0.3) with support on the interval [-Inf,Inf]."
#>
#> Prior distributions on tau (= SD of effect sizes):
#>
#> [1] "'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf]."
#> [2] "'gamma' (shape=1.5, rate=3) with support on the interval [0,Inf]."
#> [3] "'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf]."
#> [4] "'gamma' (shape=1.5, rate=3) with support on the interval [0,Inf]."
#> [5] "'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf]."
#> [6] "'gamma' (shape=1.5, rate=3) with support on the interval [0,Inf]."
plot(sensitivity, "tau", "prior")
#> Prior distributions on d (= overall effect size):
#>
#> [1] "'t' (location=0, scale=0.707, nu=1) with support on the interval [-Inf,Inf]."
#> [2] "'t' (location=0, scale=0.707, nu=1) with support on the interval [-Inf,Inf]."
#> [3] "'norm' (mean=0, sd=0.5) with support on the interval [-Inf,Inf]."
#> [4] "'norm' (mean=0, sd=0.5) with support on the interval [-Inf,Inf]."
#> [5] "'norm' (mean=0.5, sd=0.3) with support on the interval [-Inf,Inf]."
#> [6] "'norm' (mean=0.5, sd=0.3) with support on the interval [-Inf,Inf]."
#>
#> Prior distributions on tau (= SD of effect sizes):
#>
#> [1] "'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf]."
#> [2] "'gamma' (shape=1.5, rate=3) with support on the interval [0,Inf]."
#> [3] "'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf]."
#> [4] "'gamma' (shape=1.5, rate=3) with support on the interval [0,Inf]."
#> [5] "'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf]."
#> [6] "'gamma' (shape=1.5, rate=3) with support on the interval [0,Inf]."
plot(sensitivity, "tau", "posterior")
#> Prior distributions on d (= overall effect size):
#>
#> [1] "'t' (location=0, scale=0.707, nu=1) with support on the interval [-Inf,Inf]."
#> [2] "'t' (location=0, scale=0.707, nu=1) with support on the interval [-Inf,Inf]."
#> [3] "'norm' (mean=0, sd=0.5) with support on the interval [-Inf,Inf]."
#> [4] "'norm' (mean=0, sd=0.5) with support on the interval [-Inf,Inf]."
#> [5] "'norm' (mean=0.5, sd=0.3) with support on the interval [-Inf,Inf]."
#> [6] "'norm' (mean=0.5, sd=0.3) with support on the interval [-Inf,Inf]."
#>
#> Prior distributions on tau (= SD of effect sizes):
#>
#> [1] "'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf]."
#> [2] "'gamma' (shape=1.5, rate=3) with support on the interval [0,Inf]."
#> [3] "'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf]."
#> [4] "'gamma' (shape=1.5, rate=3) with support on the interval [0,Inf]."
#> [5] "'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf]."
#> [6] "'gamma' (shape=1.5, rate=3) with support on the interval [0,Inf]."
plot(sensitivity, "tau", "prior")
#> Prior distributions on d (= overall effect size):
#>
#> [1] "'t' (location=0, scale=0.707, nu=1) with support on the interval [-Inf,Inf]."
#> [2] "'t' (location=0, scale=0.707, nu=1) with support on the interval [-Inf,Inf]."
#> [3] "'norm' (mean=0, sd=0.5) with support on the interval [-Inf,Inf]."
#> [4] "'norm' (mean=0, sd=0.5) with support on the interval [-Inf,Inf]."
#> [5] "'norm' (mean=0.5, sd=0.3) with support on the interval [-Inf,Inf]."
#> [6] "'norm' (mean=0.5, sd=0.3) with support on the interval [-Inf,Inf]."
#>
#> Prior distributions on tau (= SD of effect sizes):
#>
#> [1] "'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf]."
#> [2] "'gamma' (shape=1.5, rate=3) with support on the interval [0,Inf]."
#> [3] "'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf]."
#> [4] "'gamma' (shape=1.5, rate=3) with support on the interval [0,Inf]."
#> [5] "'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf]."
#> [6] "'gamma' (shape=1.5, rate=3) with support on the interval [0,Inf]."
plot(sensitivity, "tau", "posterior")
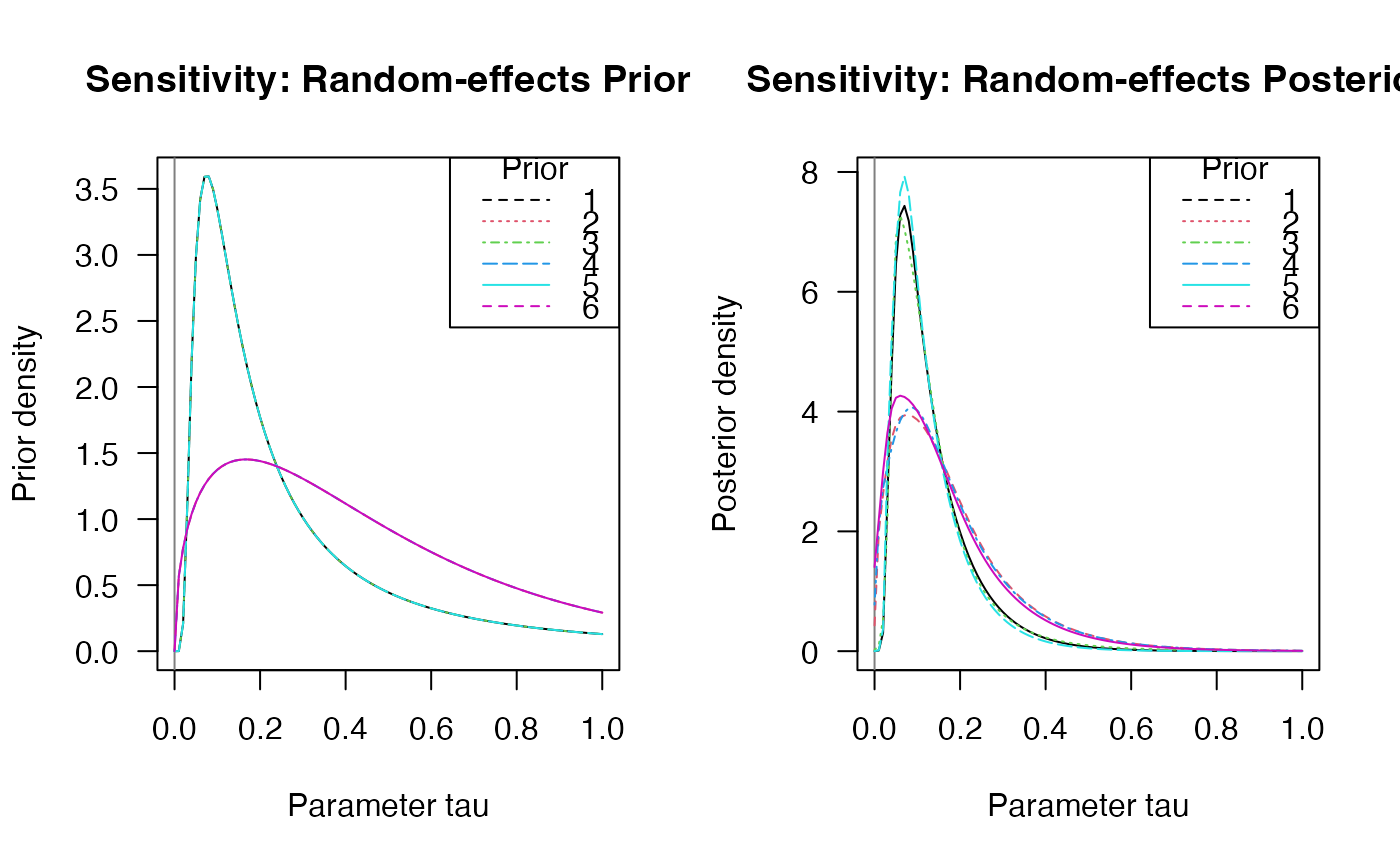 #> Prior distributions on d (= overall effect size):
#>
#> [1] "'t' (location=0, scale=0.707, nu=1) with support on the interval [-Inf,Inf]."
#> [2] "'t' (location=0, scale=0.707, nu=1) with support on the interval [-Inf,Inf]."
#> [3] "'norm' (mean=0, sd=0.5) with support on the interval [-Inf,Inf]."
#> [4] "'norm' (mean=0, sd=0.5) with support on the interval [-Inf,Inf]."
#> [5] "'norm' (mean=0.5, sd=0.3) with support on the interval [-Inf,Inf]."
#> [6] "'norm' (mean=0.5, sd=0.3) with support on the interval [-Inf,Inf]."
#>
#> Prior distributions on tau (= SD of effect sizes):
#>
#> [1] "'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf]."
#> [2] "'gamma' (shape=1.5, rate=3) with support on the interval [0,Inf]."
#> [3] "'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf]."
#> [4] "'gamma' (shape=1.5, rate=3) with support on the interval [0,Inf]."
#> [5] "'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf]."
#> [6] "'gamma' (shape=1.5, rate=3) with support on the interval [0,Inf]."
# }
#> Prior distributions on d (= overall effect size):
#>
#> [1] "'t' (location=0, scale=0.707, nu=1) with support on the interval [-Inf,Inf]."
#> [2] "'t' (location=0, scale=0.707, nu=1) with support on the interval [-Inf,Inf]."
#> [3] "'norm' (mean=0, sd=0.5) with support on the interval [-Inf,Inf]."
#> [4] "'norm' (mean=0, sd=0.5) with support on the interval [-Inf,Inf]."
#> [5] "'norm' (mean=0.5, sd=0.3) with support on the interval [-Inf,Inf]."
#> [6] "'norm' (mean=0.5, sd=0.3) with support on the interval [-Inf,Inf]."
#>
#> Prior distributions on tau (= SD of effect sizes):
#>
#> [1] "'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf]."
#> [2] "'gamma' (shape=1.5, rate=3) with support on the interval [0,Inf]."
#> [3] "'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf]."
#> [4] "'gamma' (shape=1.5, rate=3) with support on the interval [0,Inf]."
#> [5] "'invgamma' (shape=1, scale=0.15) with support on the interval [0,Inf]."
#> [6] "'gamma' (shape=1.5, rate=3) with support on the interval [0,Inf]."
# }